Applications for the CAD software extend far beyond medicine and throughout the burgeoning field of
synthetic biology, which involves redesigning organisms to give them new abilities. For example, we envision users designing solutions for biomanufacturing; it's possible that society could reduce its reliance on petroleum thanks to microorganisms that produce valuable chemicals and materials. And to aid the fight against climate change, users could design microorganisms that ingest and lock up carbon, thus reducing atmospheric carbon dioxide (the main driver of global warming).
Our consortium,
GP-write, can be understood as a sequel to the Human Genome Project, in which scientists first learned how to "read" the entire genetic sequence of human beings. GP-write aims to take the next step in genetic literacy by enabling the routine "writing" of entire genomes, each with tens of thousands of different variations. As genome writing and editing becomes more accessible, biosafety is a top priority. We're building safeguards into our system from the start to ensure that the platform isn't used to craft dangerous or pathogenic sequences.
Need a quick refresher on genetic engineering? It starts with DNA, the double-stranded molecule that encodes the instructions for all life on our planet. DNA is composed of four types of nitrogen bases—adenine (A), thymine (T), guanine (G), and cytosine (C)—and the sequence of those bases determines the biological instructions in the DNA. Those bases pair up to create what look like the rungs of a long and twisted ladder. The human genome (meaning the entire DNA sequence in each human cell) is composed of approximately 3 billion base-pairs. Within the genome are sections of DNA called genes, many of which code for the production of proteins; there are more than 20,000 genes in the human genome.
The
Human Genome Project, which produced the first draft of a human genome in 2000, took more than a decade and cost about $2.7 billion in total. Today, an individual's genome can be sequenced in a day for $600, with some predicting that the $100 genome is not far behind. The ease of genome sequencing has transformed both basic biological research and nearly all areas of medicine. For example, doctors have been able to precisely identify genomic variants that are correlated with certain types of cancer, helping them to establish screening regimens for early detection. However, the process of identifying and understanding variants that cause disease and developing targeted therapeutics is still in its infancy and remains a defining challenge.
Until now, genetic editing has been a matter of changing one or two genes within a massive genome; sophisticated techniques like
CRISPR can create targeted edits, but at a small scale. And although many software packages exist to help with gene editing and synthesis, the scope of those software algorithms is limited to single or few gene edits. Our CAD program will be the first to enable editing and design at genome-scale, allowing users to change thousands of genes, and it will operate with a degree of abstraction and automation that allows designers to think about the big picture. As users create new genome variants and study the results in cells, each variant's traits and characteristics (called its phenotype) can be noted and added to the platform's libraries. Such a shared database could vastly speed up research on complex diseases.
What's more, current genomic design software requires human experts to predict the effect of edits. In a future version, GP-write's software will include predictions of phenotype to help scientists understand if their edits will have the desired effect. All the experimental data generated by users can feed into a machine-learning program, improving its predictions in a virtuous cycle. As more researchers leverage the CAD platform and share data (the open-source platform will be freely available to academia), its predictive power will be enhanced and refined.
Our first version of the CAD software will feature a user-friendly graphical interface enabling researchers to upload a species' genome, make thousands of edits throughout the genome, and output a file that can go directly to a DNA synthesis company for manufacture. The platform will also enable design sharing, an important feature in the collaborative efforts required for large-scale genome-writing initiatives.
There are clear parallels between CAD programs for electronic and genome design. To make a gadget with four transistors, you wouldn't need the help of a computer. But today's systems may have billions of transistors and other components, and designing them would be impossible without design-automation software. Likewise, designing just a snippet of DNA can be a manual process. But sophisticated genomic design—with thousands to tens of thousands of edits across a genome—is simply not feasible without something like the CAD program we're developing. Users must be able to input high-level directives that are executed across the genome in a matter of seconds.
Our CAD program will be the first to enable editing at genome-scale, with a degree of abstraction and automation that allows designers to think about the big picture.
A good CAD program for electronics includes certain design rules to prevent a user from spending a lot of time on a design, only to discover that it can't be built. For example, a good program won't let the user put down transistors in patterns that can't be manufactured or put in a logic that doesn't make sense. We want the same sort of design-for-manufacture rules for our genomic CAD program. Ultimately, our system will alert users if they're creating sequences that can't be manufactured by synthesis companies, which currently have limitations such as trouble with certain repetitive DNA sequences. It will also inform users if their biological logic is faulty; for example, if the gene sequence they added to code for the production of a protein won't work, because they've mistakenly included a "stop production" signal halfway through.
But other aspects of our enterprise seem unique. For one thing, our users may import huge files containing billions of base-pairs. The genome of the
Polychaos dubium, a freshwater amoeboid, clocks in at 670 billion base-pairs—that's over 200 times larger than the human genome! As our CAD program will be hosted on the cloud and run on any Internet browser, we need to think about efficiency in the user experience. We don't want a user to click the "save" button and then wait ten minutes for results. We may employ the technique of lazy loading, in which the program only uploads the portion of the genome that the user is working on, or implement other tricks with caching.
Getting a DNA sequence into the CAD program is just the first step, because the sequence, on its own, doesn't tell you much. What's needed is another layer of annotation to indicate the structure and function of that sequence. For example, a gene that codes for the production of a protein is composed of three regions: the promoter that turns the gene on, the coding region that contains instructions for synthesizing RNA (the next step in protein production), and the termination sequence that indicates the end of the gene. Within the coding region, there are "exons," which are directly translated into the amino acids that make up proteins and "introns," intervening sequences of nucleotides that are removed during the process of gene expression. There are existing standards for this annotation that we want to improve on, so our standardized interface language will be readily interpretable by people all over the world.
 The CAD program from GP-write will enable users to apply high-level directives to edit a genome, including inserting, deleting, modifying, and replacing certain parts of the sequence. GP-write
The CAD program from GP-write will enable users to apply high-level directives to edit a genome, including inserting, deleting, modifying, and replacing certain parts of the sequence. GP-write
Once a user imports the genome, the editing engine will enable the user to make changes throughout the genome. Right now, we're exploring different ways to efficiently make these changes and keep track of them. One idea is an approach we call genome algebra, which is analogous to the algebra we all learned in school. In mathematics, if you want to get from the number 1 to the number 10, there are infinite ways to do it. You could add 1 million and then subtract almost all of it, or you could get there by repeatedly adding tiny amounts. In algebra, you have a set of operations, costs for each of those operations, and tools that help organize everything.
In genome algebra, we have four operations: we can insert, delete, invert, or edit sequences of nucleotides. The CAD program can execute these operations based on certain rules of genomics, without the user having to get into the details. Similar to the "
PEMDAS rule" that defines the order of operations in arithmetic, the genome editing engine must order the user's operations correctly to get the desired outcome. The software could also compare sequences against each other, essentially checking their math to determine similarities and differences in the resulting genomes.
In a later version of the software, we'll also have algorithms that advise users on how best to create the genomes they have in mind. Some altered genomes can most efficiently be produced by creating the DNA sequence from scratch, while others are more suited to large-scale edits of an existing genome. Users will be able to input their design objectives and get recommendations on whether to use a synthesis or editing strategy—or a combination of the two.
 Users can import any genome (here, the E. coli bacteria genome), and create many edited versions; the CAD program will automatically annotate each version to show the changes made. GP-write
Users can import any genome (here, the E. coli bacteria genome), and create many edited versions; the CAD program will automatically annotate each version to show the changes made. GP-write
Our goal is to make the CAD program a "one-stop shop" for users, with the help of the members of our Industry Advisory Board: Agilent Technologies, a global leader in life sciences, diagnostics and applied chemical markets; the DNA synthesis companies Ansa Biotechnologies, DNA Script, and Twist Bioscience; and the gene editing automation companies Inscripta and Lattice Automation. (Lattice was founded by coauthor Douglas Densmore). We are also partnering with biofoudries such as the Edinburgh Genome Foundry that can take synthetic DNA fragments, assemble them, and validate them before the genome is sent to a lab for testing in cells.
Users can most readily benefit from our connections to DNA synthesis companies; when possible, we'll use these companies' APIs to allow CAD users to place orders and send their sequences off to be synthesized. (In the case of DNA Script, when a user places an order it would be quickly printed on the company's DNA printers; some dedicated users might even buy their own printers for more rapid turnaround.) In the future, we'd like to make the ordering step even more user-friendly by suggesting the company best suited to the manufacture of a particular sequence, or perhaps by creating a marketplace where the user can see prices from multiple manufacturers, the way people do on airfare sites.
We've recently added two new members to our Industrial Advisory Board, each of which brings interesting new capabilities to our users.
Catalog Technologies is the first commercially viable platform to use synthetic DNA for massive digital storage and computation, and could eventually help users store vast amounts of genomic data generated on GP-write software. The other new board member is SOSV's IndieBio, the leader in biotech startup development. It will work with GP-write to select, fund, and launch companies advancing genome-writing science from IndieBio's New York office. Naturally, all those startups will have access to our CAD software.
We're motivated by a desire to make genome editing and synthesis more accessible than ever before. Imagine if high-school kids who don't have access to a wet lab could find their way to genetic research via a computer in their school library; this scenario could enable outreach to future genome design engineers and could lead to a more diverse workforce. Our CAD program could also entice people with engineering or computational backgrounds—but with no knowledge of biology—to contribute their skills to genetic research.
Because of this new level of accessibility, biosafety is a top priority. We're planning to build several different levels of safety checks into our system. There will be user authentication, so we'll know who's using our technology. We'll have biosecurity checks upon the import and export of any sequence, basing our "prohibited" list on the standards devised by the
International Gene Synthesis Consortium (IGSC), and updated in accordance with their evolving database of pathogens and potentially dangerous sequences. In addition to hard checkpoints that prevent a user from moving forward with something dangerous, we may also develop a softer system of warnings.
Imagine if high-school kids who don't have access to a lab could find their way to genetic research via a computer in their school library.
We'll also keep a permanent record of redesigned genomes for tracing and tracking purposes. This record will serve as a unique identifier for each new genome and will enable proper attribution to further encourage sharing and collaboration. The goal is to create a broadly accessible resource for researchers, philanthropies, pharmaceutical companies, and funders to share their designs and lessons learned, helping all of them identify fruitful pathways for advancing R&D on genetic diseases and environmental health. We believe that the authentication of users and annotated tracking of their designs will serve two complementary goals: It will enhance biosecurity while also engendering a safer environment for collaborative exchange by creating a record for attribution.
One project that will put the CAD program to the test is a grand challenge adopted by GP-write, the Ultra-Safe Cell Project. This effort, led by coauthor Farren Isaacs and Harvard professor George Church, aims to create a human cell line that is resistant to viral infection. Such virus-resistant cells could be a huge boon to the biomanufacturing and pharmaceutical industry by enabling the production of more robust and stable products, potentially driving down the cost of biomanufacturing and passing along the savings to patients.
The Ultra-Safe Cell Project relies on a technique called recoding. To build proteins, cells use combinations of three DNA bases, called codons, to code for each amino acid building block. For example, the triplet 'GGC' represents the amino acid glycine, TTA represents leucine, GTC represents valine, and so on. Because there are 64 possible codons but only 20 amino acids, many of the codons are redundant. For example, four different codons can code for glycine: GGT, GGC, GGA, and GGG. If you replaced a redundant codon in all genes (or 'recode' the genes), the human cell could still make all of its proteins. But viruses—whose genes would still include the redundant codons and which rely on the host cell to replicate—would not be able to translate their genes into proteins. Think of a key that no longer fits into the lock; viruses trying to replicate would be unable to do so in the cells' machinery, rendering the recoded cells virus-resistant.
This concept of recoding for viral resistance has already been demonstrated. Isaacs, Church, and their colleagues reported in a 2013 paper in
Science that, by removing all 321 instances of a single codon from the genome of the E. coli bacterium, they could impart resistance to viruses which use that codon. But the ultra-safe cell line requires edits on a much grander scale. We estimate that it would entail thousands to tens of thousands of edits across the human genome (for example, removing specific redundant codons from all 20,000 human genes). Such an ambitious undertaking can only be achieved with the help of the CAD program, which can automate much of the drudge work and let researchers focus on high-level design.
The famed physicist
Richard Feynman once said, "What I cannot create, I do not understand." With our CAD program, we hope geneticists become creators who understand life on an entirely new level.
From Your Site Articles
Related Articles Around the Web




 DNA, the molecule that encodes instructions for life, is composed of four types of nitrogen bases, which pair up to create what look like the rungs of a twisted ladder.James Provost
DNA, the molecule that encodes instructions for life, is composed of four types of nitrogen bases, which pair up to create what look like the rungs of a twisted ladder.James Provost
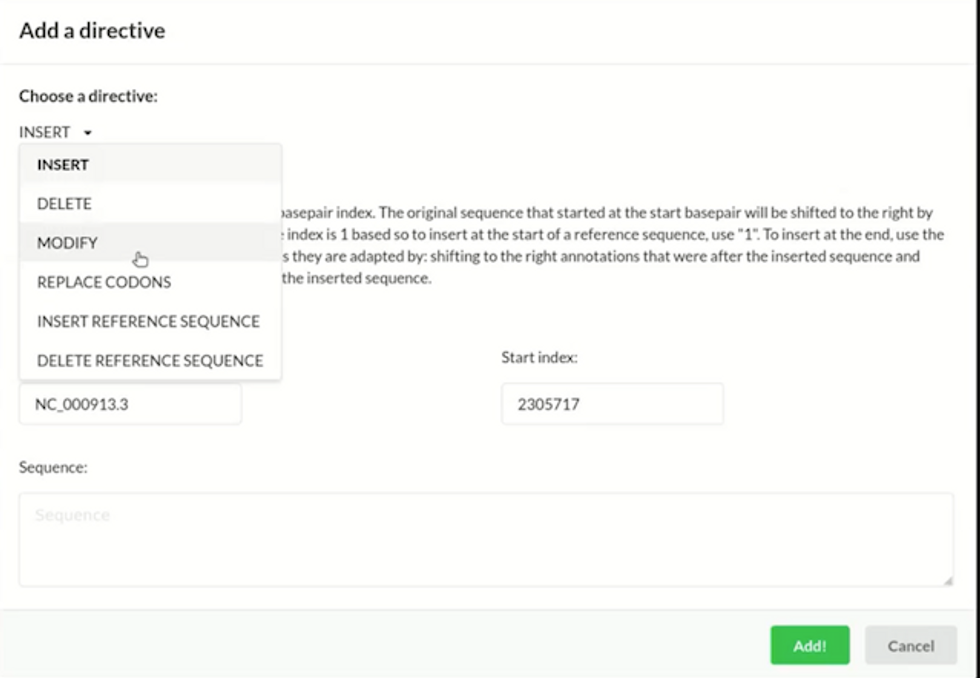 The CAD program from GP-write will enable users to apply high-level directives to edit a genome, including inserting, deleting, modifying, and replacing certain parts of the sequence. GP-write
The CAD program from GP-write will enable users to apply high-level directives to edit a genome, including inserting, deleting, modifying, and replacing certain parts of the sequence. GP-write
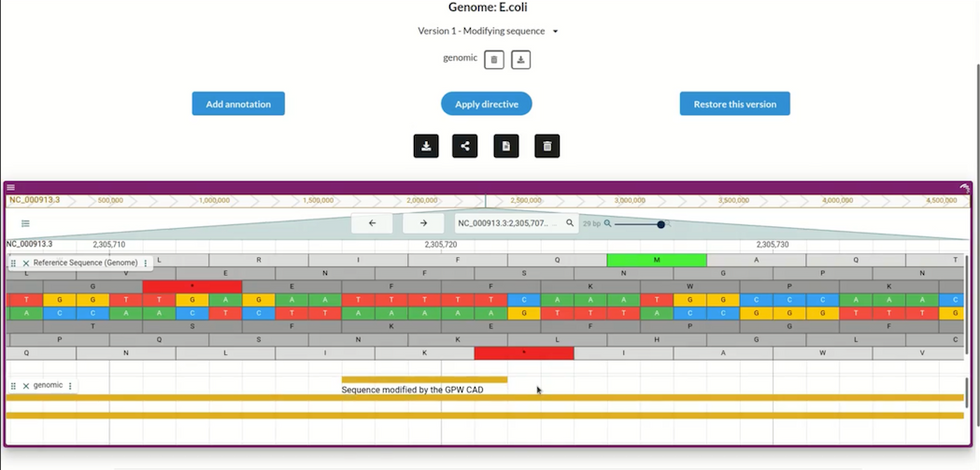 Users can import any genome (here, the E. coli bacteria genome), and create many edited versions; the CAD program will automatically annotate each version to show the changes made. GP-write
Users can import any genome (here, the E. coli bacteria genome), and create many edited versions; the CAD program will automatically annotate each version to show the changes made. GP-write